
SmartRank
User manual
Version ${project.version}
Welcome to the manual for SmartRank version ${project.version}.
SmartRank is a robust likelihood ratio software that enables searching of national DNA-databases with complex DNA-profiles. The methodology is optimized to enable searches of (increasingly) voluminous DNA-databases, while keeping the rate of false negative and false positive errors at a minimum. SmartRank enables a more efficient searching of DNA-databases with partial and/or complex profiles. SmartRank should primarily be used as a tool to acquire intelligence data.
qDesignationShutdown option. The effects of the Q Designation Shutdown are described in
ISHI 2016 proceedings located in the 'manual' subfolder of the SmartRank installation folder.
The first step of using SmartRank is to load the database containing the candidate profiles. The database can be loaded on the DNA database tab. Any format problems will be displayed in the table at the bottom of the panel. Details of which specimen, locus and cause of the problem can be viewed here. Continuing with format problems still present will result in the problematic locus or specimen - depending on the nature of the problem - being excluded from the analysis.
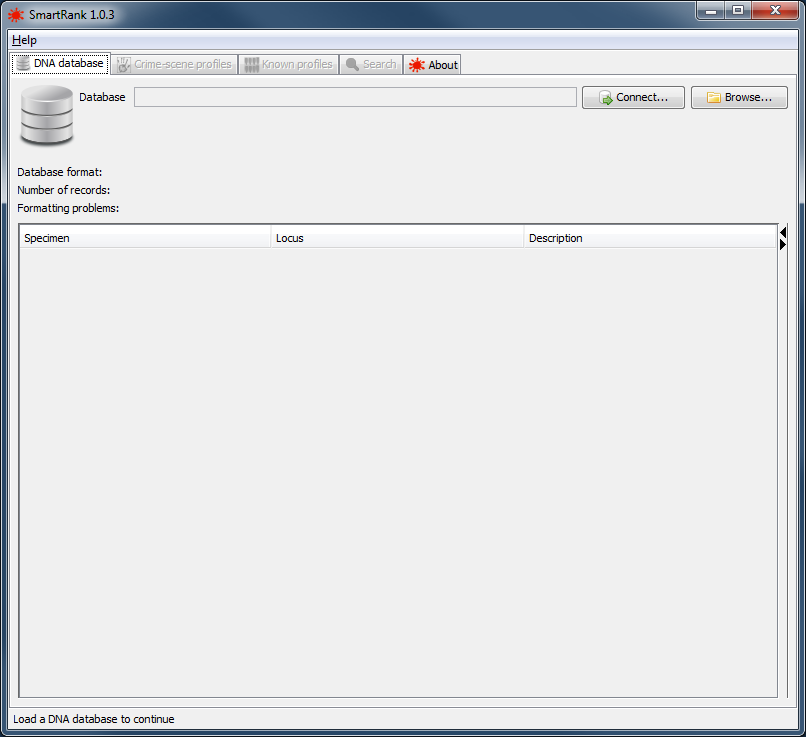 |
the database tab |
The DNA database can be read from a file or from a direct database connection.
CODIS users, please see DatabaseExport for the SQL query can be used to export the specimens to a suitable format.
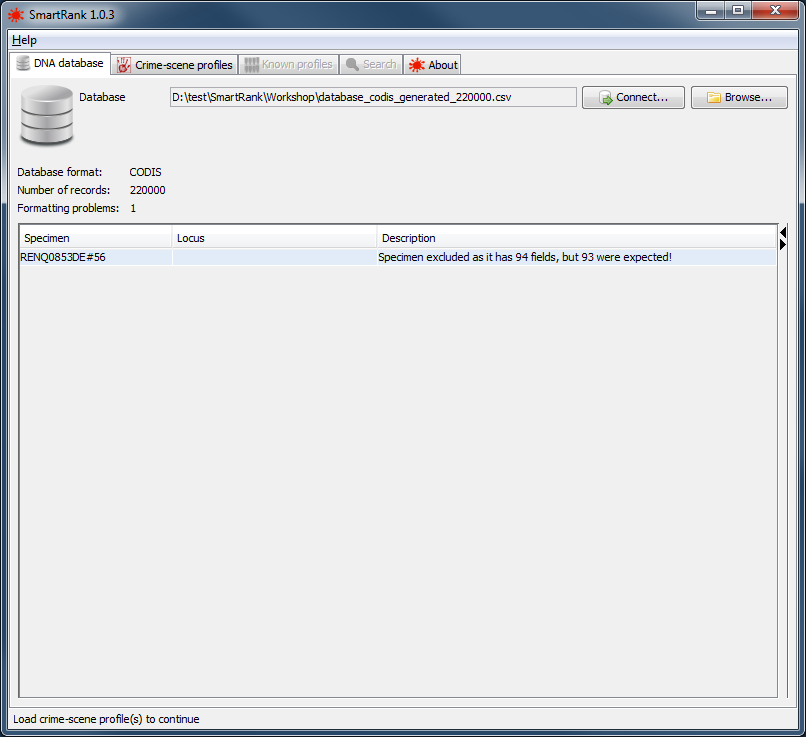 |
a successfully loaded database with a single formatting error |
SmartRank has the capability to connect directly Microsoft SQLServer, SAP Adaptive Server Enterprise or H2 databases. On the Database tab, click the Connect... button to bring up the Database Settings screen on which you can review or configure the connection to the database.
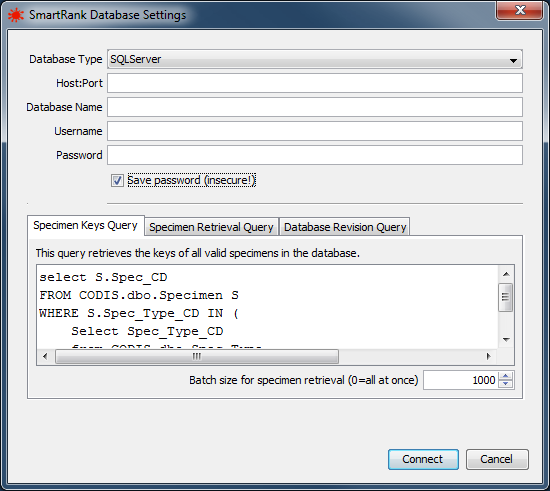 |
the Database Settings dialog |
| Setting | Description |
|---|---|
| Database Type | The type of DBMS that is used for the DNA database. Supported types are SQLServer, Sybase and H2. |
| Host:Port | The network address and port of the database server. |
| Database Name | The schema name of the DNA database. |
| Username | The username under which to log in. |
| Password | The password for logging in to the database. |
| Save Password | Indicates whether the password is to be stored in the configuration file. IMPORTANT NOTE: the storage of the password is in no way secure! Only use this option if the machine SmartRank runs on is in a trusted environment. |
| Specimen Keys Query | The query to be executed in order to obtain a list of all eligible specimens in the database. |
| Specimen Retrieval Batch Size | This value indicates how many specimens are to be retrieved from the database in one go. This can be set to 0 to retrieve all specimens, but this may have adverse effects on the responsiveness of SmartRank while specimens are being retrieved! |
| Specimen Retrieval Query | The query to be executed in order to obtain the data for some or all of the specimens in the database. |
| Database Revision Query | The query to be executed in order to obtain a value that uniquely identifies the current state of the database. |
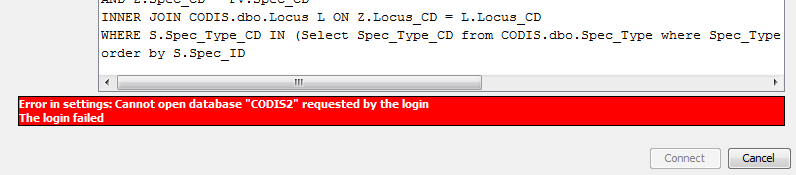 |
example of an error message in the Database Settings dialog |
| SpecimenId | locus | allele |
|---|---|---|
| ID-1 | vWA | 11 |
| ID-1 | vWA | 12 |
| ID-1 | FGA | 13.2 |
| ID-1 | FGA | 14 |
| ID-2 | vWA | 8 |
| ID-2 | vWA | 12 |
| ID-2 | FGA | 17 |
| ID-2 | FGA | 18 |
| SpecimenId | locus | allele | Specimen Type |
|---|---|---|---|
| ID-1 | vWA | 11 | Convicted Offender |
| ID-1 | vWA | 12 | Convicted Offender |
| ID-1 | FGA | 13.2 | Convicted Offender |
| ID-1 | FGA | 14 | Convicted Offender |
| ID-2 | vWA | 8 | Missing Person |
| ID-2 | vWA | 12 | Missing Person |
| ID-2 | FGA | 17 | Missing Person |
| ID-2 | FGA | 18 | Missing Person |
| ID-3 | vWA | 8 | Convicted Offender |
| ID-3 | vWA | 12 | Convicted Offender |
| ID-3 | FGA | 17 | Convicted Offender |
| ID-3 | FGA | 18 | Convicted Offender |
| Specimen Type | |
|---|---|
| Convicted Offender | 2 |
| Missing Person | 1 |
| SpecimenId | locus | allele |
|---|---|---|
| ID-1 | vWA | 11 12 |
| ID-1 | FGA | 13.2 14 |
| ID-2 | vWA | 8 12 |
| ID-2 | FGA | 17 18 |
| SpecimenId | locus | allele |
|---|---|---|
| ID-1 | vWA | 11 |
| ID-2 | vWA | 8 |
| ID-1 | vWA | 12 |
| ID-2 | vWA | 12 |
| ID-1 | FGA | 13.2 |
| ID-2 | FGA | 17 |
| ID-1 | FGA | 14 |
| ID-2 | FGA | 18 |
| RecordName | Marker | PCR Value |
|---|---|---|
| ID-1 | vWA | 11 12 |
| ID-1 | FGA | 13.2 14 |
| ID-2 | vWA | 8 12 |
| ID-2 | FGA | 17 18 |
Load the mix profile(s) for crime stains on the Crime-scene profiles tab by clicking the Add button and selecting the appropriate file. After loading the sample file, the bottom table will show all loci detected in the file, along with their according alleles. In the top table, the basic information of the sample is shown along with a possibility to enable or disable the profiles for the analysis. If the file contains replicates, all these will be shown as individual samples. It is also possible to load replicates from multiple files.
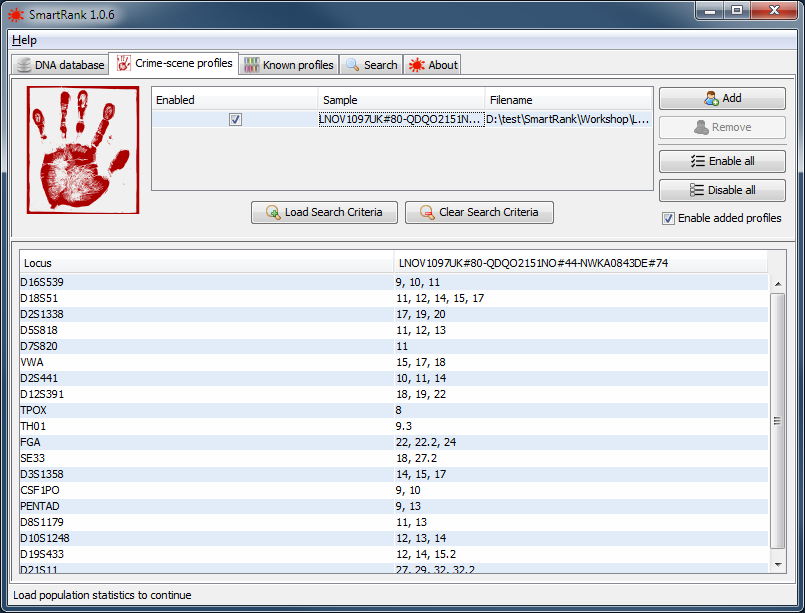 |
a successfully loaded crime scene sample. |
The mix profile file must be in tab- or comma separated format. See ProfileFormat for more information.
To allow integration with other tools, it is possible to import search settings (samples, profiles, hypotheses etc.) from a single file. The format of this file is described in SearchCriteriaFormat. Click the 'Load Search Criteria' button to select a file from which to load the search criteria. Note that all current settings are discarded and reset to their defaults before loading the file.
The 'Clear Search Criteria' button resets all settings (except those relating to the database and population statistics) to their default values. In situations where multiple independent searches are to be carried out sequentially, this guarantees a clean starting point for new searches and avoids having to restart SmartRank and re-validate the database before each search.
Optionally, just like loading crime scene profiles, you can load any known contributors to the mixed profile in the known profiles tab.
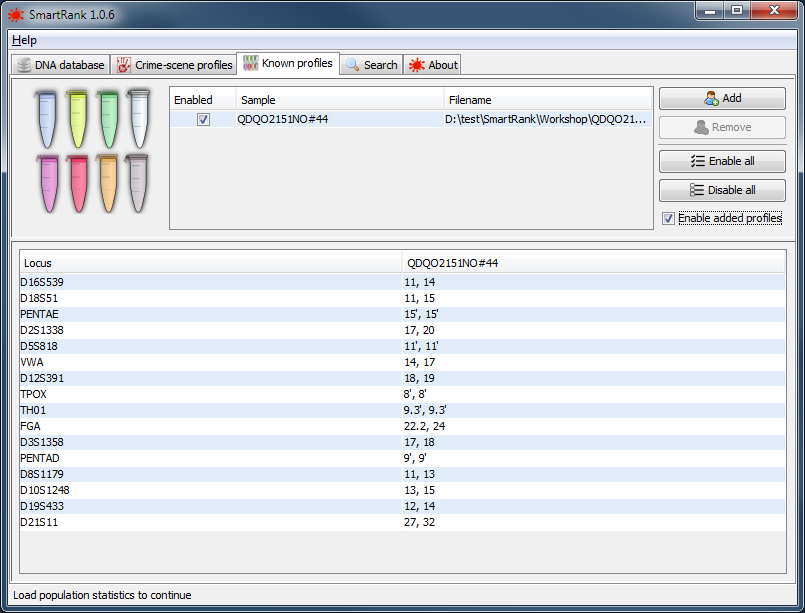 |
a successfully loaded known sample. |
The known profile file must be in tab- or comma separated format. See Profile Format for more information.
On the top half of the search tab, the hypotheses for the prosecution and defense can be formulated. Select the contributors for each hypotheses (known from the previously loaded known profiles and/or unknown by choosing the number of unknown contributors) and set their dropout probabilities. The dropout probabilities can also be estimated by SmartRank by pressing the 'Estimate dropout probabilities' button after loading the population statistics. In this case, SmartRank will use the 95% percentile of a Monte-Carlo simulation as the drop-out estimate.
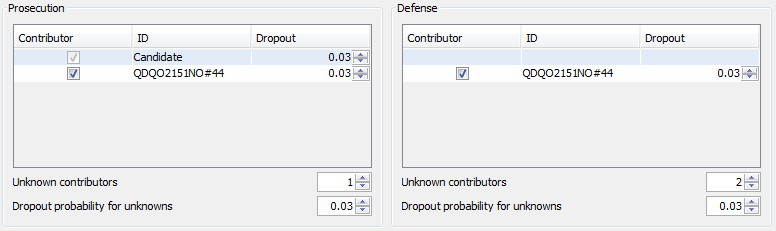 |
formulating hypotheses for prosecution and defense |
In the Parameters section of the Search tab, various settings can be configured that can influence the results of the search. See below for the possible settings and their effects.
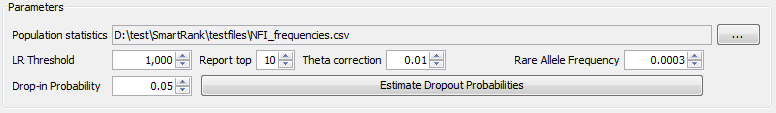 |
the Parameters section |
| Setting | Description |
|---|---|
| Population Statistics | Click the ... button to select a poulation statistics file. The population statistics file must be in tab- or comma separated format. See PopulationFormat for more information. |
| LR Threshold | Specimens that result in an LR at or below this threshold will not be reported. |
| Report top | At most this number of specimens will be reported. |
| Theta correction | The value for the Theta correction. To see how this parameter influences the search, please refer to the statistical specifications located in the 'manual' subfolder of the SmartRank installation folder. |
| Rare Allele Frequency | This frequency will be assigned to alleles that are not present in the population statistics. |
| Dropin Probability | The probability that an unrelated allele dropped into the mixture. To see how this parameter influences the search, please refer to the statistical specifications. |
| Estimate Dropout Probabilities | Click this button to perform a dropout estimation. Please refer to the statistical specifications for more information. This feature can be disabled in the restrictions file (see PresettingParameters) |
When both hypotheses have been formulated and the parameters have been set, the search can be started by pressing the 'Search' button. The estimated time to complete the search will be shown and once it is completed, any results will be displayed in the results box at the bottom half of the screen. If necessary, the search can be aborted at any time by pressing the red cross next to the progress bar.
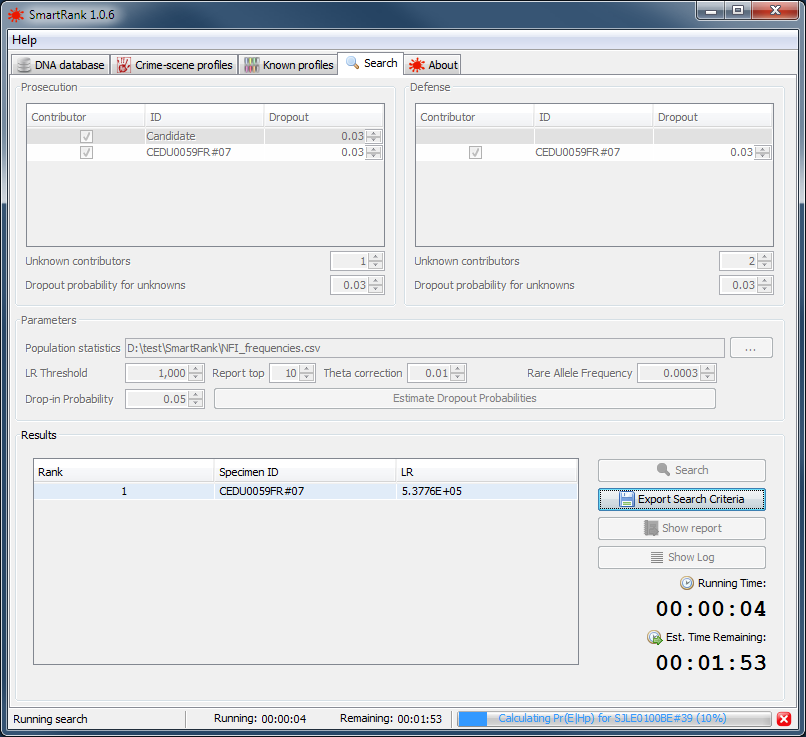 |
SmartRank running a search |
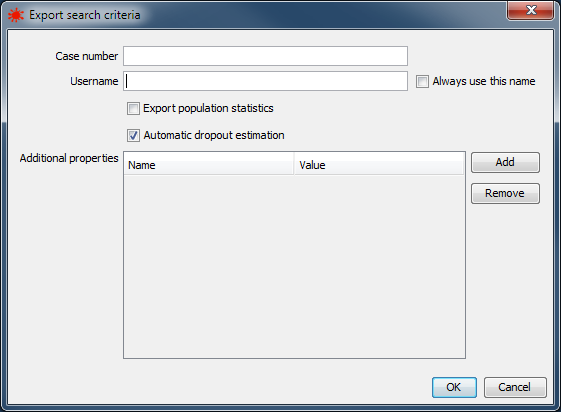 |
Search criteria export dialog |
After the search is completed, the results (if any) will be displayed in the results panel at the bottom half of the screen. Any results exceeding the preset LR threshold will be displayed here. The Show Report button opens the PDF containing the search results. Pressing the Control key while clicking this button will open the folder containing the report file instead of the file itself. The Show Log button open the log file generated during the search. Here also, pressing the Control key while clicking this button will open the folder containing the log file.
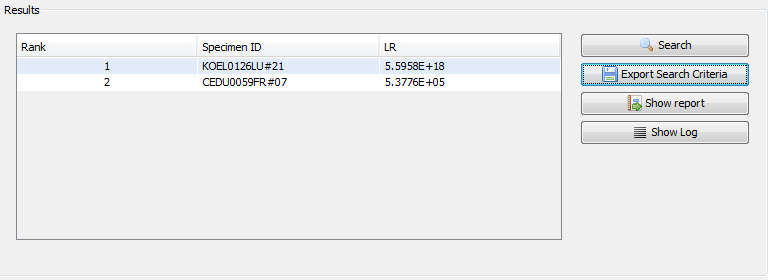 |
displaying of search results |
The details of a matching profile can be viewed by double clicking the profile in the results table. In this detailed view, allelic mismatches between the sample and the candidate are highlighted in red.
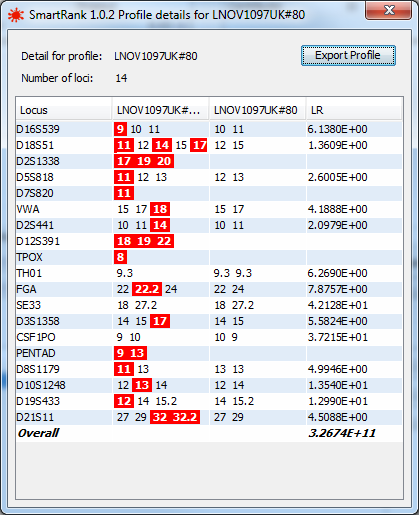 |
more details of a single result |
Whenever a search is performed, a logfile is created recording the parameters with which the search is conducted, the input files used and the results obtained. This logfile is a plain text file that can be opened in any text editor. By default, the logfile is placed in a subfolder named SmartRankResults that is created in the folder containing the crimescene samples. The default location and file name of the logfile can be configured in the restrictions file (see PresettingParameters).
After a search is performed, a report in PDF format is created containing the parameters and results of the search. By default, the report is placed in a subfolder named SmartRankResults that is created in the folder containing the crimescene samples. The default location and file name of the report can be configured in the restrictions file (see PresettingParameters).
Next to the logfile and report, SmartRank will export the reported profiles in CSV format in the same location as the report. The CSV files are in a format that is suitable to be read by LRmixStudio. If this is not desired, the feature can be disabled in the restrictions file (see PresettingParameters).
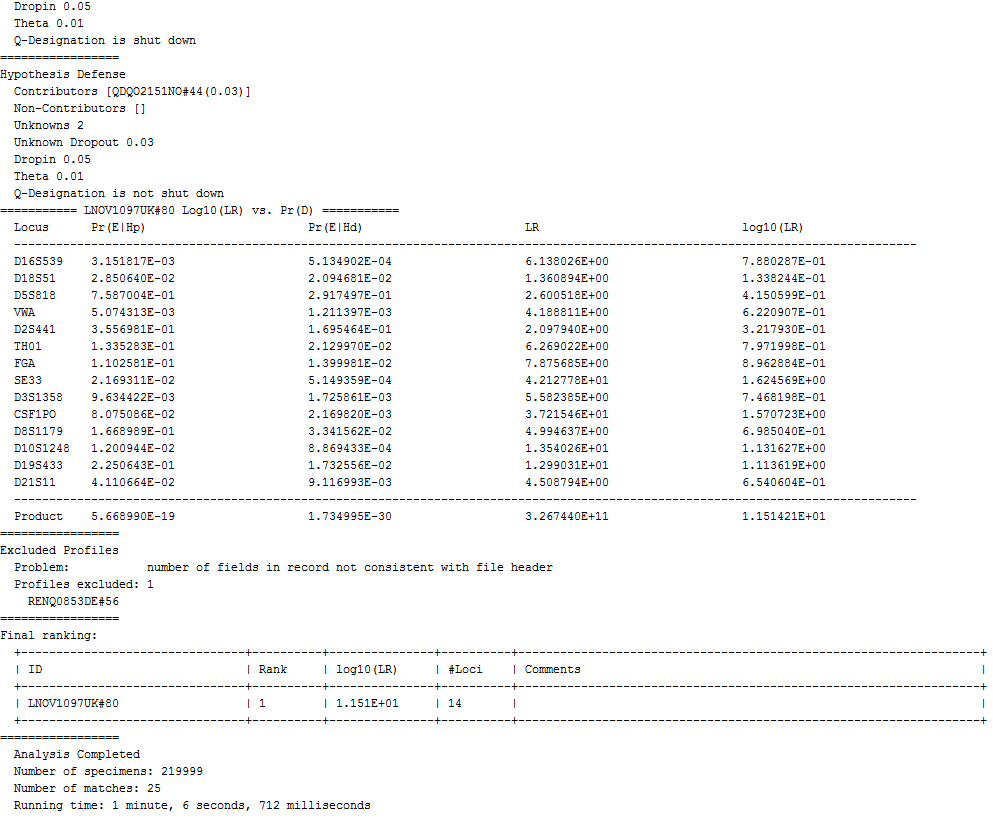 |
excerpt of the search log |
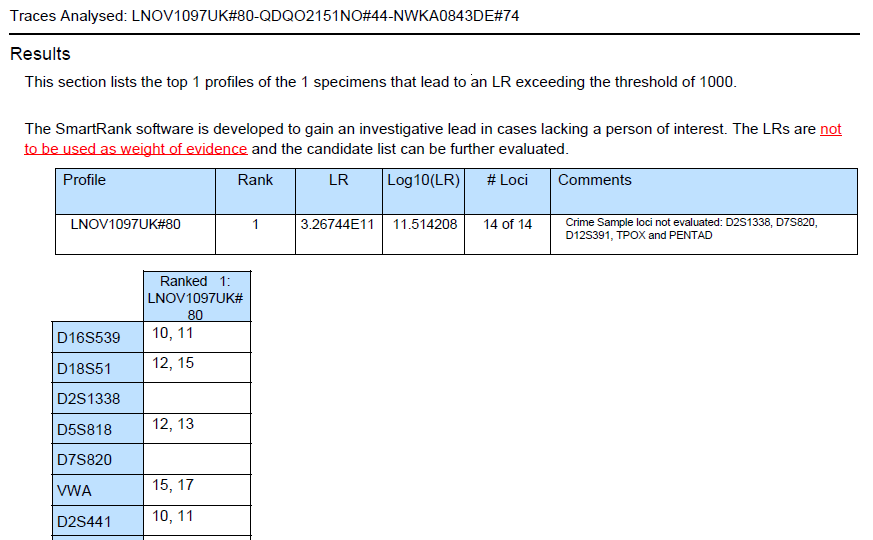 |
excerpt of the search report |
SmartRank uses a number of files to store configuration data. To support concurrently running instances of SmartRank each using their own configuration, the names of these files can be configured. The files are:
| Property | Function | Allowed values |
|---|---|---|
| windowTitle | Sets a string that will be included after the application name and version in the title of the SmartRank window. Can be useful to distinguish between concurrently running instances of SmartRank. | Any string. This property is optional and can be omitted, in which case the window title will be SmartRank ${project.version}. |
| Property | Function | Allowed values |
|---|---|---|
| automaticParameterEstimationEnabled | Makes SmartRank automatically estimate dropout probabilities using a Monte Carlo simulation. Manual input of dropout probabilities will be disabled. | true/false |
| batchMode | When set to true, SmartRank will start in batch mode (see BatchMode). | true/false |
| caseLogFilename | Sets the name of the logfile that is generated for each search. | A template describing the name of case log files. See TemplateStrings for more information. |
| dropoutMinimum | Sets a minimum value for drop-out probability. | 0.0 to 0.99 |
| dropoutDefault | Sets a default value for drop-out probability. | 0.0 to 0.99 |
| dropoutMaximum | Sets a maximum value for drop-out probability. | 0.0 to 0.99 |
| dropinMinimum | Sets a minimum value for drop-in probability. | 0.0 to 0.99 |
| dropinDefault | Sets a default value for drop-in probability. | 0.0 to 0.99 |
| dropinMaximum | Sets a maximum value for drop-in probability. | 0.0 to 0.99 |
| thetaMinimum | Sets a minimum value for theta. | 0.0 to 0.99 |
| thetaDefault | Sets a default value for theta. | 0.0 to 0.99 |
| thetaMaximum | Sets a maximum value for theta. | 0.0 to 0.99 |
| maximumStoredResults | Sets a maximum number of returned likelihood ratios. | Any positive number, 250 by default |
| maximumPathLength | Sets the maximum length of paths for logfiles and reports. This to prevent creating files that cannot be read on some versions of Windows. | Any positive number, 128 by default |
| maximumUnknownCount | Sets a maximum number of unknown contributors. | Any positive number, 4 by default |
| minimumNumberOfLoci | Sets a minimum number of loci for a profile to be included in an analysis. | Any positive number, 1 by default |
| interactiveParameterEstimationEnabled | Makes dropout probability estimation manually executable. | true/false |
| parameterEstimationIterations | The number of iterations used for estimating the dropout probability. | Any positive number, 10000 by default |
| reportFileName | Sets the path, where analysis reports are exported to. | A template describing the name of analysis report files. See TemplateStrings for more information. |
| reportTemplateFileName | Sets the path to the template, used for creating analysis reports. | A filepath |
| outputRootFolder | When importing search criteria from a file, the output path specified in the file - if any - will be resolved relative to this folder. Especially useful when the machine generating the XML has different drive mappings than the machine running SmartRank. | A filepath |
| exportMatchingProfilesAfterSearch | Indicates whether the profiles with LRs exceeding the LR Threshold should be exported to csv files after a search. | true/false |
| defaultLRThreshold | The default value for the LR threshold. | Any positive integer, 1000 by default |
| qDesignationShutdown | Indicates whether the Q Designation should be shut down for the calculation of Pr(E|Hp). Please note that changing this value is not recommended, as it changes the numerical results of the program, and can cause performance problems. The setting is only present for testing purposes. |
true/false, true by default |
| windowCloseBlockedInBatchMode | Indicates whether closing the application window is allowed when SmartRank Batch Mode is in running state. If set to true, closing application window while SmartRank is in running state (i.e. the run button has been clicked) will minimize the window instead. | true/false, false by default |
| batchMode.autoStart | When in batch mode, this option can be specified to automatically connect to the database or load a database file and start the batch process when SmartRank is started. | db automatically connect to the database and start batch processing.file automatically load the last-loaded file and start batch processing. |
| batchMode.retentiondays | When in batch mode, this option can be specified to automatically remove jobs that are older than the specified number of days. | a positive integer. Specifying 0 will clean up processed jobs immediately. Default value is 14. |
Some values in the restrictions file are specified in Template Strings. These are strings containing placeholder markers that are replaced with actual values to form the final string. Placeholders are enclosed in '{' and '}' characters (e.g. {DATE}, {TIME}) This is useful for situations where the requested value is not fixed, but depends on the input at runtime.
Example: the template string The current time is {TIME} will be resolved to The time is 133000 if the current system time is half past one in the afternoon.
The following placeholders are defined:
Batch mode was introduced to reduce the number of manual actions when processing a large number of search queries. To activate Batch Mode, set the batchMode property to 'true' in the restrictions file (see PresettingParameters).
When started in batch mode - and after loading a database - the Batch Mode tab is enabled. On this tab a directory can be selected that will be monitored for Search Criteria files. The detected files will be shown in a list, and some basic management of search jobs is available.
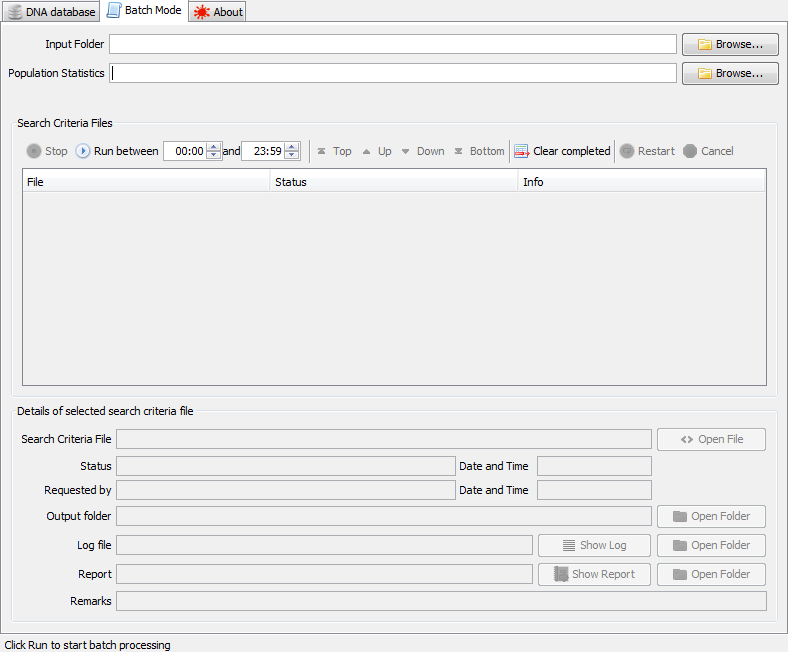 |
the Batch Mode tab |
Batch Mode constantly monitors a configured input folder for search criteria files. The path of this directory can be typed into the Input Folder box, or the Browse button can be used to select a directory using a file chooser dialog.
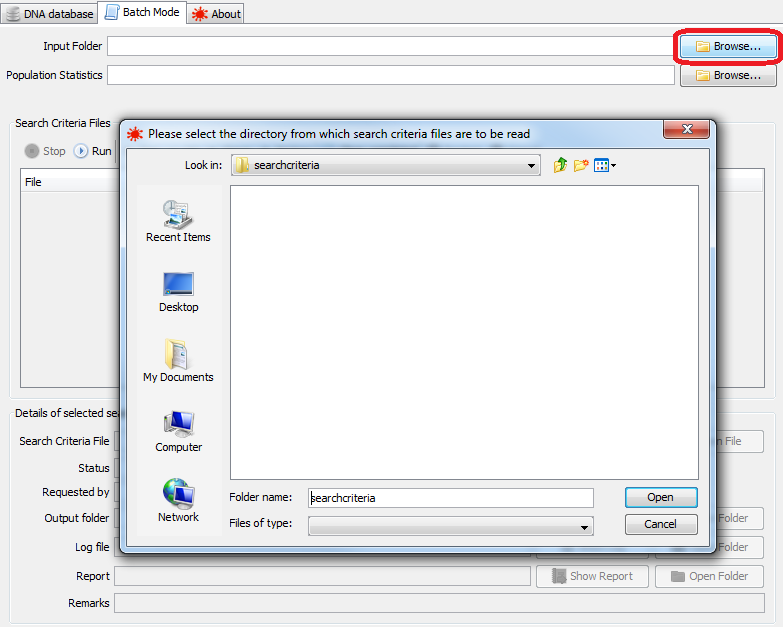 |
selecting a directory to scan for search criteria files |
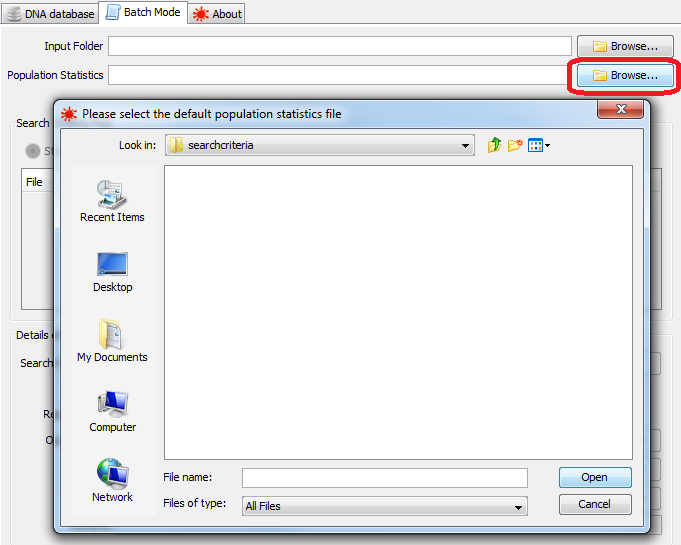 |
selecting a default population statistics file |
The batch mode tab will list the filename, status and any remarks for all search criteria files found in the input directory. This directory is periodically scanned for new files, which will be added to the displayed list automatically. Select a line in the table to see more details in the lower section:
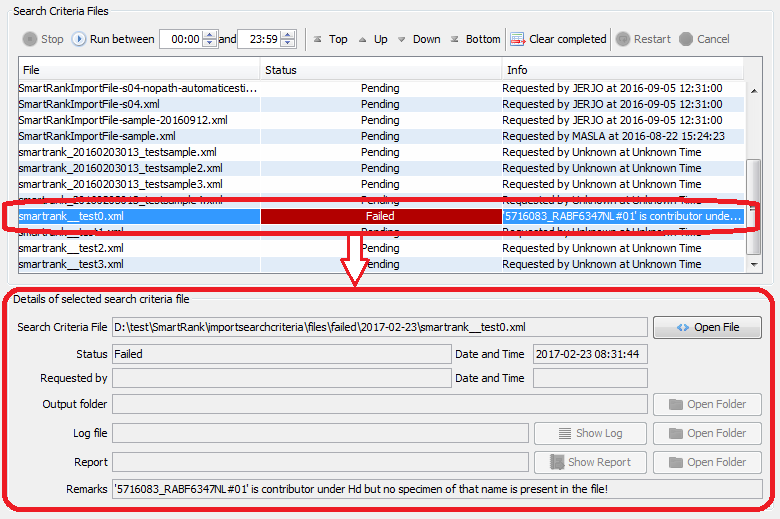 |
showing details of a selected file. In this case a file where the sample data for a contributor is listed under Hd could not be found. |
| Status | Description | File Location |
|---|---|---|
| Pending | Indicates that SmartRank has detected the presence of the file, and has verified that the file contains search criteria. | These files are present in the selected directory |
| Processing | Indicates that a file is currently being processed. | Before being processed, files are moved into the 'processing' subdirectory. If this subdirectory does not exist, it is created. |
| Failed | Indicates that an error was encountered when reading or processing the file. Depending on the nature of the failure, the files may be rescheduled by selecting the file and clicking the Restart button. | These files are moved to the 'failed' subdirectory. If this subdirectory does not exist, it is created. |
| Succeeded | Indicates that a search was performed using the criteria from the file, and that a report and logfile were created. Note that this does not necessarily mean that any matches were obtained. | These files are moved to the 'succeeded' subdirectory. If this subdirectory does not exist, it is created. |
| Interrupted | Indicates that the user interrupted the search before all specimens in the database were evaluated. Interrupted files can be re-scheduled by selecting them and clicking the Restart button. | These files are moved from the 'processing' subdirectory back to the of the input directory. |
| Cancelled | Indicates that the this file should be skipped. These files can be re-scheduled by selecting them and clicking the Restart button. | These files are moved to the 'Failed' directory. |
Above the list of files a toolbar is displayed with which a number of actions can be performed.
 |
the toolbar |
| Button | Description |
|---|---|
| Stop | Stops processing files. If a file is currently being processed, SmartRank waits until this has completed. This button is disabled until the Run button is clicked. |
| Run | Starts performing searches for the files in the input directory. Files are processed in the order in which they appear in the list. The Up/Down/Top/Bottom buttons can be used to prioritized searches. Note that if the windowCloseBlockedInBatchMode option is set in the Restrictions file, the SmartRank window cannot be closed while Batch Mode is in running state. |
| Start Time | This field contains a time in 24-hour notation. SmartRank will wait until this time to start processing search criteria files after the Run button is clicked. Use the up and down arrows to increase or decrease the time, or type a value in the edit box. The default value for this field is 00:00. |
| End Time | This field contains a time in 24-hour notation. SmartRank stop processing search criteria files after this time. If a file is being processed when the End Time is reached, this search is completed but no subsequent file will be picked up until the Start Time is once again reached. Use the up and down arrows to increase or decrease the time, or type a value in the edit box. The default value for this field is 23:59. |
| Top | Moves the selected file to the top of the list. |
| Up | Moves the selected file up one position. |
| Down | Moves the selected file down one position. |
| Bottom | Moves the selected file to the bottom of the list. |
| Clear completed | Removes Failed, Cancelled and Successful files from the list. |
| Restart | Sets the status of the selected file to Pending. This button is only enabled if a file with status Failed, Cancelled or Interrupted is selected. |
| Cancel | Sets the status of the currently selected file to Cancelled. |
| Edit PPScript | Opens a script editor and allows editing and testing of the post-processing script. |
When all files have been processed, SmartRank will continue to monitor the selected directory, and automatically process any new search criteria files, until the Stop button is clicked or SmartRank is closed.
SmartRank can perform post-processing steps after every search job. These steps are configured in JavaScript. Please note that the post-processing script is run after both successful and failed jobs. It is up to the script to determine whether specific actions need to be taken for either of these cases. Clicking the Edit PPScript button in the toolbar opens up the post-processing script editor.
 |
the post-processing script editor |
console.log statements.| Button | Description |
|---|---|
| Save | Saves the contents of the script to a new file. |
| Load | Loads a script from a file. |
| Test | Executes the script in test mode. Test settings are loaded from the SearchCriteriaForTest.xml file located in the scriptingtestfiles folder of the application directory. In test mode, the file copyToDir method supplied by the FileUtils object will not actually copy a file, but rather log a descriptive text to the console window. |
job helper object represents the current search job. It has functions to access the job's attributes such as whether the job finished successfully,
the name of the search criteria file, the search result etc. Some of the available functions are listed here:
| Function | Parameters | Description |
|---|---|---|
| getFileName | Returns a string containing the absolute filename of the search criteria file. | |
| isSucceeded | returns true if the job was run successfully, false otherwise. | |
| getErrorMessage | If the job failed, this function returns a description of the error. | |
| getLogFileName | If available, this function returns the name of the job logfile. | |
| getReportFileName | If available, this function returns the name of the job's report file. | |
| getThreshold | returns the LR threshold. LRs below this threshold will not be reported. | |
| getNumberOfLRsOverThreshold | returns the number of profiles that yielded an LR exceeding the configured threshold. If the job failed, 0 is returned. | |
| getDuration | This function returns the duration in milliseconds of the search. | |
| getSearchResults | Returns an object describing the search results. See the information panel in the code editor for this object's properties and functions. |
log helper object allows a script to write log lines into the application log.
| Function | Parameters | Description |
|---|---|---|
| info | String msg | Writes the supplied string to the application log at info level. |
| warn | String msg | Writes the supplied string to the application log at warning level. |
| error | String msg | Writes the supplied string to the application log at error level. |
FileUtils helper object allows a script to perform limited file manipulation operations.
| Function | Parameters | Description |
|---|---|---|
| fileName | String path | Parses the supplied absolute path and returns only the file's name and extension. |
| copyToDir | String fileName, String outputDir | Copies a file to a directory. On Windows, both the standard drive:\path format and UNC format (e.g. \\host\path) can be used. If a file of the same name already exists in the output directory, the new file's name is changed to make it unique. |
Check your Java version by opening a command prompt and typing:
java -version
SmartRank requires Java 1.7.0 or higher.
The following text may be present in the Comments section of the Results table in the SmartRank report.
Specimen loci not evaluated: TH01 and VWA
The names of the loci in question will vary according to your specific situation.
SmartRank only calculates an LR for the loci that are present in all of the following locations:
Crime Sample loci not evaluated: TH01 and VWA
When you see a dialog that looks something like this:
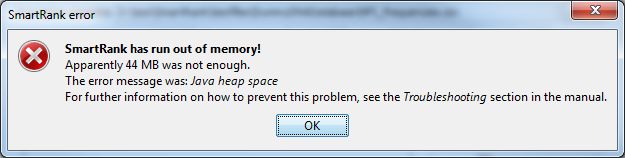
it means
that SmartRank does not have enough memory to continue.
java -jar smartrank-${project.version}.jar -Xmx2048m -Xms2048mUsing the query below, a SmartRank-compatible database file can be exported from a CODIS SQLServer database.
SELECT specimenId, CSF1PO_1, CSF1PO_2, CSF1PO_3, CSF1PO_4, D10S1248_1, D10S1248_2, D10S1248_3, D10S1248_4, D12S391_1, D12S391_2, D12S391_3, D12S391_4, D13S317_1, D13S317_2, D13S317_3, D13S317_4, D16S539_1, D16S539_2, D16S539_3, D16S539_4, D18S51_1, D18S51_2, D18S51_3, D18S51_4, D19S433_1, D19S433_2, D19S433_3, D19S433_4, D1S1656_1, D1S1656_2, D1S1656_3, D1S1656_4, D21S11_1, D21S11_2, D21S11_3, D21S11_4, D22S1045_1, D22S1045_2, D22S1045_3, D22S1045_4, D2S1338_1, D2S1338_2, D2S1338_3, D2S1338_4, D2S441_1, D2S441_2, D2S441_3, D2S441_4, D3S1358_1, D3S1358_2, D3S1358_3, D3S1358_4, D5S818_1, D5S818_2, D5S818_3, D5S818_4, D7S820_1, D7S820_2, D7S820_3, D7S820_4, D8S1179_1, D8S1179_2, D8S1179_3, D8S1179_4, F13A1_1, F13A1_2, F13A1_3, F13A1_4, FES_FPS_1, FES_FPS_2, FES_FPS_3, FES_FPS_4, FGA_1, FGA_2, FGA_3, FGA_4, [Penta D_1], [Penta D_2], [Penta D_3], [Penta D_4], [Penta E_1], [Penta E_2], [Penta E_3], [Penta E_4], SE33_1, SE33_2, SE33_3, SE33_4, TH01_1, TH01_2, TH01_3, TH01_4, TPOX_1, TPOX_2, TPOX_3,TPOX_4, vWA_1, vWA_2, vWA_3, vWA_4 FROM ( SELECT S.Spec_ID AS [specimenId], CAST(rtrim(Locus) AS VARCHAR(50)) + '_' + CAST(PV.Band_Num AS VARCHAR(2)) [locus], rtrim(PV.PCR_Value) AS [data] FROM CODIS.dbo.Specimen S INNER JOIN CODIS.dbo.Sizing Z ON S.Spec_CD = Z.Spec_CD INNER JOIN CODIS.dbo.PCR_Value PV ON Z.Locus_CD = PV.Locus_CD AND Z.Spec_CD = PV.Spec_CD INNER JOIN CODIS.dbo.Locus L ON Z.Locus_CD = L.Locus_CD WHERE S.Spec_Type_CD IN ( SELECT Spec_Type_CD FROM CODIS.dbo.Spec_Type WHERE Spec_Type IN ( 'Convicted Offender', 'Suspect, Known', 'Elimination, Known' ) ) ) x PIVOT ( MAX(DATA) FOR locus IN ( CSF1PO_1, D10S1248_1, D12S391_1, D13S317_1, D16S539_1, D18S51_1, D19S433_1, D1S1656_1, D21S11_1, D22S1045_1, D2S1338_1, D2S441_1, D3S1358_1, D5S818_1, D7S820_1, D8S1179_1, F13A1_1, FES_FPS_1, FGA_1, [Penta D_1], [Penta E_1], SE33_1, TH01_1, TPOX_1, vWA_1, CSF1PO_2, D10S1248_2, D12S391_2, D13S317_2, D16S539_2, D18S51_2, D19S433_2, D1S1656_2, D21S11_2, D22S1045_2, D2S1338_2, D2S441_2, D3S1358_2, D5S818_2, D7S820_2, D8S1179_2, F13A1_2, FES_FPS_2, FGA_2, [Penta D_2], [Penta E_2], SE33_2, TH01_2, TPOX_2, vWA_2, CSF1PO_3, D10S1248_3, D12S391_3, D13S317_3, D16S539_3, D18S51_3, D19S433_3, D1S1656_3, D21S11_3, D22S1045_3, D2S1338_3, D2S441_3, D3S1358_3, D5S818_3, D7S820_3, D8S1179_3, F13A1_3, FES_FPS_3, FGA_3, [Penta D_3], [Penta E_3], SE33_3, TH01_3, TPOX_3, vWA_3, CSF1PO_4, D10S1248_4, D12S391_4, D13S317_4, D16S539_4, D18S51_4, D19S433_4, D1S1656_4, D21S11_4, D22S1045_4, D2S1338_4, D2S441_4, D3S1358_4, D5S818_4, D7S820_4, D8S1179_4, F13A1_4, FES_FPS_4, FGA_4, [Penta D_4], [Penta E_4], SE33_4, TH01_4, TPOX_4, vWA_4 ) ) AS p
Currently, the only accepted format for a DNA database is a CODIS CSV format. A CODIS CSV file is a comma-separated file listing allelic information for one or multiple specimens. The first line contains a header; starting with specimen ID, followed by identifiers for genetic markers (e.g. THO1 or D3S1358). Specimens are listed below this header with a unique identifier and alleles for the above mentioned genetic markers.
Below is an example of a DNA database file, containing allelic information of 10 different specimens.
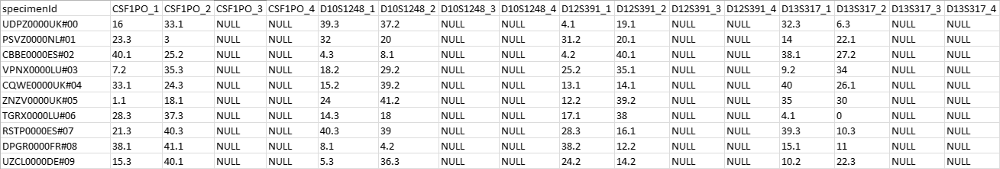
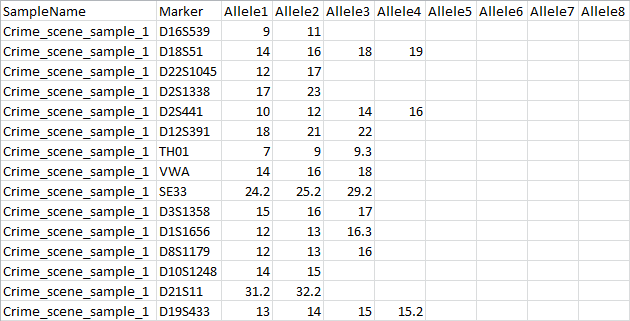
The accepted formats for profile files (crime scene- or known) are tab- or comma-separated files. The first line of the file contains a preset header of the profiles; it lists a sample name, a marker and up to eight alleles. Every detected marker in the sample is listed per row, describing what sample it originates from, its identifier and the allelic information associated with that marker.
Below is an example of a crime scene sample file, containing allelic information of 15 identified markers. Known profile files use the same format.
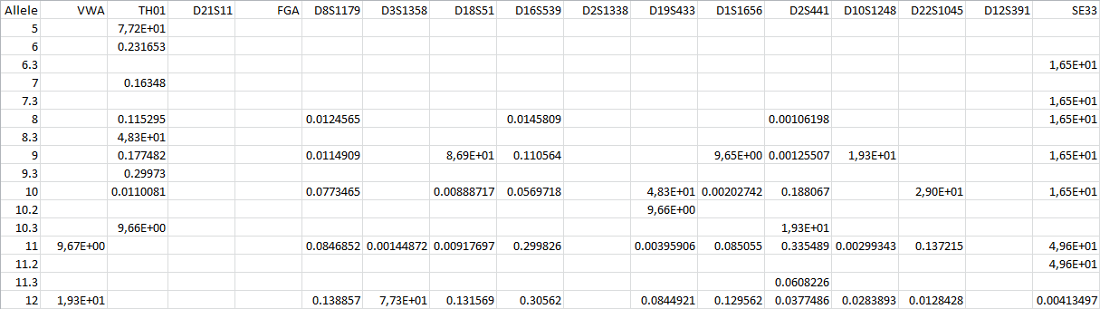
The accepted format for population statistics files is tab- or comma-separated. Alleles are displayed vertically, loci horizontally. The cells where alleles and loci cross displays the probability of existence of that allele in that specific locus in the population. Should the allele never appear in a locus, the cell remains blank.
Below is an example of a population statistics file, showing allelic probabilities of 16 different loci.
Search criteria can be imported from an XML or JSON file. An example of search criteria in XML format is shown below. See the xsd folder of the SmartRank installation folder for an XSD file describing the Search Criteria file format.
<SmartRankImportFile version="1.0" xmlns="urn:SmartRankImportFile-schema">
<!-- The case folder is the place where logfiles and reports are written to.
This overrides the location configured in the restrictions file -->
<CaseFolder>2016/01 januari/2016.01.01.001</CaseFolder>
<!-- The user id of the person who generated this file -->
<userid>MASLA</userid>
<!-- The date and time that the file was generated (not used in smartrank, but present for tracability reasons) -->
<dateTime>2016-08-22 15:24:23</dateTime>
<!-- This contains references to all replicates (crime-scene profiles) -->
<Replicates>
<!-- Each replicate get a Replicate item here. The name refers to the name of a Specimen (see below) -->
<Replicate name="CEDU0059FR#07-KOEL0126LU#21"/>
</Replicates>
<!-- The value to use for dropin -->
<Dropin>0.10</Dropin>
<!-- The value to use for theta -->
<Theta>0.03</Theta>
<!-- Specification of Hp. The candidateDropout attribute defines the dropout for the candidate sample. -->
<HP candidateDropout="0.10">
<!-- Contains references to the samples that contributors under Hp, and their corresponding dropout -->
<Contributors>
<!-- Each sample that is a contributor under Hp gets a Contributor item here.
The name refers to the name of a Specimen (see below) -->
<Contributor name="CEDU0059FR#07" dropout="0.11"/>
</Contributors>
<!-- Contains the settings for unknown contributors under Hp -->
<Unknowns count="1" dropout="0.12"/>
</HP>
<!-- Specification of Hd. -->
<HD>
<!-- Contains references to the samples that contributors under Hd, and their corresponding dropout -->
<Contributors>
<!-- Each sample that is a contributor under Hd gets a Contributor item here.
The name refers to the name of a Specimen (see below) -->
<Contributor name="CEDU0059FR#07" dropout="0.13"/>
</Contributors>
<!-- Contains the settings for unknown contributors under Hd -->
<Unknowns count="2" dropout="0.14"/>
</HD>
<!-- For every loaded crime-scene profile and known profile, a Specimen entry
is included listing the contents of the sample. -->
<Specimen name="CEDU0059FR#07-KOEL0126LU#21">
<!-- Every locus in the sample gets a Locus item here -->
<Locus name="D16S539">
<!-- Every allele in this locus gets an Allele item here -->
<Allele value="11"/>
<Allele value="12"/>
<Allele value="13"/>
</Locus>
<Locus name="D18S51">
<Allele value="14"/>
<Allele value="15"/>
</Locus>
</Specimen>
<Specimen name="CEDU0059FR#07">
<Locus name="D16S539">
<Allele value="11"/>
<Allele value="11"/>
</Locus>
<Locus name="D18S51">
<Allele value="14"/>
<Allele value="15"/>
</Locus>
</Specimen>
</SmartRankImportFile>
Below is an example of search criteria in JSON format
{
"caseFolder": "2016/01 januari/2016.01.01.001",
"userid": "MASLA",
"dateTime": "2016-08-22 15:24:23",
"replicates": {
"replicate": [
{
"name": "CEDU0059FR#07-KOEL0126LU#21"
}
]
},
"dropin": 0.10,
"theta": 0.03,
"lrThreshold": 1234,
"maximumNumberOfResults": 10,
"rareAlleleFrequency": 0.000239808153477218,
"hp": {
"contributors": {
"contributor": [
{
"name": "CEDU0059FR#07",
"dropout": "0.11"
}
]
},
"nonContributors": null,
"unknowns": {
"count": 1,
"dropout": "0.12"
},
"candidateDropout": "0.10"
},
"hd": {
"contributors": {
"contributor": [
{
"name": "CEDU0059FR#07",
"dropout": "0.13"
}
]
},
"nonContributors": null,
"unknowns": {
"count": 2,
"dropout": "0.14"
}
},
"specimen": [
{
"locus": [
{
"allele": [
{
"value": "11"
},
{
"value": "12"
}
],
"name": "D16S539"
},
{
"allele": [
{
"value": "14"
},
{
"value": "15"
}
],
"name": "D18S51"
}
],
"name": "CEDU0059FR#07-KOEL0126LU#21"
},
{
"locus": [
{
"allele": [
{
"value": "11"
},
{
"value": "12"
}
],
"name": "D16S539"
},
{
"allele": [
{
"value": "14"
},
{
"value": "15"
}
],
"name": "D18S51"
}
],
"name": "KOEL0126LU#21"
},
{
"locus": [
{
"allele": [
{
"value": "11"
},
{
"value": "12"
}
],
"name": "D16S539"
},
{
"allele": [
{
"value": "14"
},
{
"value": "15"
}
],
"name": "D18S51"
}
],
"name": "CEDU0059FR#07"
}
],
"properties": {
"property": [
{
"name": "property1",
"value": "value1"
},
{
"name": "property2",
"value": "value2"
}
]
},
"version": 1.0
}
The following queries can be used when accessing a CODIS database directly:
select S.Spec_CD
FROM CODIS.dbo.Specimen S
WHERE S.Spec_Type_CD IN (
Select Spec_Type_CD
from CODIS.dbo.Spec_Type
where Spec_Type IN ('Convicted Offender','Suspect, Known', 'Elimination, Known'))
order by Spec_CD
SELECT S.Spec_ID as [specimenId], L.Locus, rtrim(PV.PCR_Value) as [allele]
FROM CODIS.dbo.Specimen S, CODIS.dbo.Locus L, CODIS.dbo.Sizing Z, CODIS.dbo.PCR_Value PV
WHERE S.Spec_CD = Z.Spec_CD
AND Z.Spec_CD = PV.Spec_CD
AND Z.Locus_CD = L.Locus_CD
AND Z.Locus_CD = PV.Locus_CD
AND S.Spec_CD between ? and ?
AND S.Spec_Type_CD IN (
Select Spec_Type_CD
from CODIS.dbo.Spec_Type
where Spec_Type IN ('Convicted Offender','Suspect, Known', 'Elimination, Known'))
group by s.spec_id, L.locus, PV.PCR_Value
select max(Audit_CD) FROM CODIS.dbo.Audit_Trail